
How do things really work?
Janina Sprenger, Postdoc at Lund University, is specialized in protein crystallography. Janina got in contact with Lund University through an Erasmus project in 2008, which was a part of her master in biochemistry at the University of Potsdam in Germany. In Lund, she got introduced to structural biology and methods for crystallizing proteins. Later on, she continued her PhD at the Medical Faculty at Lund University focusing on how to develop medication against diseases like malaria.
– I guess I was always interested in science and trying to understand how things really work, but also to create new functions of whatever I find in my surroundings – which wasn’t always a pleasure for my parents.
After her PhD Janina joined Sara Linse’s research group at the Biochemistry and Structural Biology department at Lund University where she is now involved in several research project. One of these has been funded within MAX4ESSFUN.
Figuring out structures of proteins
X-rays can be used to give structural information of biomolecules that allow us to understand their functions in the cell or how to cure diseases.
– In my research proteins become quickly the molecules to focus on because they are the working horses in our body, they allow us to store memory, to move, to see and they run our metabolism. Many diseases originate from malfunctions of proteins. For example Alzheimer’s disease is caused by proteins aggregating in the brain and cancer is often a result of defect proteins.
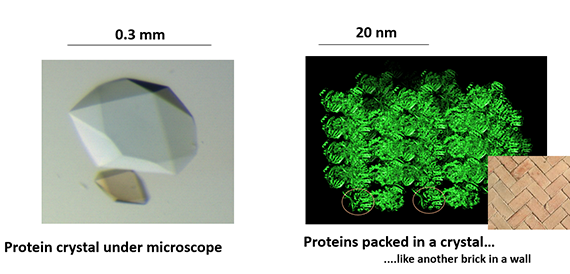
Nowadays, X-ray crystallography is the most used method to obtain atomic resolution structures of proteins. This method requires that the proteins can form a crystal.
– You may think of a brick tower where all bricks – protein molecules – are well ordered packed together to form “the building” or crystal. A protein crystal, however, is penetrated by water channels and 60-70 % of the crystal is water. These crystals are then exposed to X-rays and with help of smart computational methods we manage to get a protein structure which is an atomic model of the protein we crystallized. Several proteins, however, do not crystallize and their structure cannot be solved by X-rays.
Janina explains how a crystal host system can be used to overcome that problem.
– Instead of crystallizing these proteins we trap them inside the water channels of an already existing crystal of another protein that has very large channels or pores. We call this protein crystal host system where the porous crystals of the protein, in our case “TrpR” provide the host crystals in which we want to incorporate the “crystal guest”, the proteins which structure we want to study. We have many indications that we can bring the proteins inside the crystals, but the biggest challenge is that we need to order the proteins inside in certain direction. We are testing within this Interreg-experiment different tricks to do that.
Neutrons – a complementary method
Janina is already quite experienced in using X-rays and has conducted several experiments at different synchrotron facilities such as: ESRF in Grenoble, DESY in Hamburg (both parts of the Interreg-project), Diamond Light Source in UK and MAX-lab in Lund. In the future she believes that a neutron light source like the ESS could be very usable for her research.
– It would be even better if we could use both techniques in this project. The way of doing an experiment with neutrons is a little bit different and will take a little more time, but it would be an excellent complementary method.
Janina is sure that she will continue using research facilities such as ESS and MAX IV in the future.
– My research depends on having access to such facilities! Even if I would not continue with the protein crystal host systems, crystallography is a central technique that I am using in different projects.
Interdisciplinary and international research
– I like the interdisciplinary research which doesn´t necessarily need to be directly applied. I feel very much home in the interface between structural biology, the biomedical field and also somewhat having engineering in mind to further develop methods. For me it is a perfect combination to try to gain a deep understanding of how the human body works on a molecular level, getting hints on how living organisms evolve but also how we researchers can actually help to fight diseases.
The Interreg-experiment mainly focuses on developing new methods to study protein structures.
– It may take a long time to develop this protein crystal host system that we are working on. It may be that this system can be applied even better in different areas like material science. We still hope to make a contribution to understand the structure of some proteins that could not be crystalized yet. A lot of diseases are related to malfunction of proteins. Hopefully many diseases can be solved through learning more about the protein structure.
Janina´s overall research projects includes experts from Lund University, MAX IV and USA. She explains that the international team is important because the partners have complementary skills that they put into the project.
– Due to the cross-border set up between Lund University and MAX IV I was able to get input on the crystallography part of our project and advices how to conduct and evaluate the experiments. I also get insights into the status of the BioMAX beamline at MAX IV and can follow the process.
Janina participated in the annual MAX4ESSFUN meeting and the “Synchrotron and Neutron Scattering” Interreg workshop in October 2016. She thinks these kinds of activities are very valuable.
– The scattering workshop and the annual meeting gave me the possibility to gain expert input from different fields. Since my project is interdisciplinary it is otherwise difficult to get that kind of feedback and discussions. Also I managed to get in contact with a group in Copenhagen with similar interests which opens new possibilities of funding in the future.
Interview: Kristina Sandberg Hrbinic
Short facts:
Young researcher: Janina Sprenger
Experiment: Trapped in the crystal: Towards a new method to obtain structural information of small proteins through X-ray crystallography
Experiment period: 2016-03-01 – 2016-08-31
Supervisor: Sara Snogerup Linse, Lund University
Co-supervisor: Marjolein Thunnissen, MAX IV